Cell-free glycoprotein synthesis provides cheaper, more rapid way to make medicines
EVANSTON, Ill. — Engineering cellular biology — minus the actual cell — is a growing area of interest in biotechnology and synthetic biology. It’s known as cell-free protein synthesis, or CFPS, and it has potential to provide sustainable ways to make chemicals, medicines and biomaterials.
One major challenge stands in the way: the ability to manufacture glycosylated proteins, or proteins with a carbohydrate attachment. Glycosylation is crucial for a wide range of important biological processes, and the ability to understand and control this mechanism is vital for disease treatment and prevention.
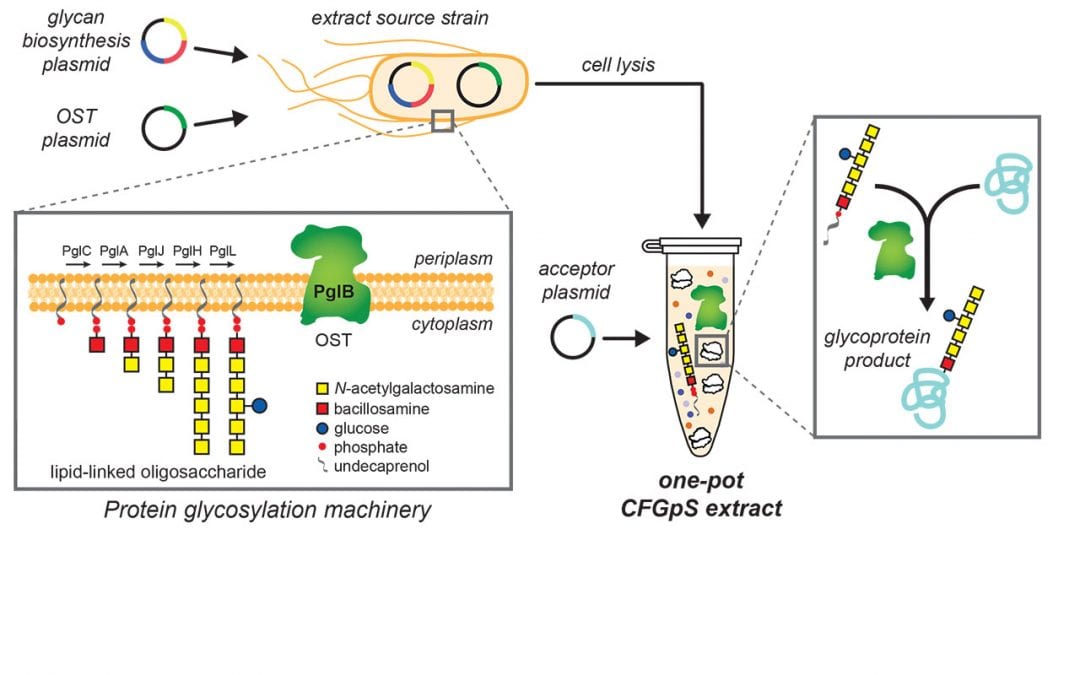
Chemistry of Life Processes resident faculty member Michael Jewett, the Charles Deering McCormick Professor of Teaching Excellence and associate professor of chemical and biological engineering at Northwestern University, and Matthew DeLisa, the William L. Lewis Professor of Engineering in the Smith School of Chemical and Biomolecular Engineering at Cornell University, have teamed up to develop a novel approach that overcomes this challenge. Their first-of-its-kind system leverages recent advances in CFPS while adding the crucial glycosylation component in a simplified, “one-pot” reaction. In the future, their cell-free glycoprotein synthesis system could be freeze-dried and reactivated for point-of-use protein synthesis by simply adding water.
“If you really want to have a useful, portable and deployable vaccine or therapeutic protein technology that’s cell free, you have to figure out the carbohydrate attachment,” DeLisa said. “That’s, in essence, what we’ve done in a very powerful way.”
Jewett and DeLisa are co-senior authors of “Single-pot Glycoprotein Biosynthesis Using a Cell-Free Transcription-Translation System Enriched with Glycosylation Machinery,” published today (July 12) in Nature Communications. Thapakorn Jaroentomeechai, a Ph.D. student in the DeLisa Research Group, and Jessica Stark, a Ph.D. student and National Science Foundation Graduate Fellow in the Jewett group, are co-first authors.
“What’s so exciting is that this collaboration is the perfect combination of enabling technologies from our labs,” Jewett said.
DeLisa studies the molecular mechanisms underlying protein biogenesis in the complex environment of a living cell, such as Escherichia coli (E. coli). While his lab has made some notable breakthroughs, the limitations of this area, he said, are the cell walls themselves.
An expert in cell-free synthetic biology, Jewett avoids the cell walls altogether by removing the biomachinery from the confines of the cell. “In bacterial cell engineering, you’re constantly in a tug of war,” said Jewett, who is co-director of Northwestern’s Center for Synthetic Biology. “You’re introducing a mechanism or capability that’s of interest to you as a scientist, but what the cell is trying to do for itself is grow and survive.”
For their new method, Jewett and DeLisa’s teams prepared cell extracts from an optimized laboratory strain of E. coli that were selectively enriched with key glycosylation components. The resulting extracts enabled a simplified reaction scheme, which the team has dubbed cell-free glycoprotein synthesis (CFGpS).
“A major advance of this work is that our cell-free extracts contain all of the molecular machinery for protein synthesis and protein glycosylation,” Stark said. “What that means is you only need to add DNA instructions for your protein of interest to make a glycoprotein in CFGpS. This is a drastic simplification from cell-based methods and allows us to make sophisticated glycoprotein molecules in less than a day.”
And the CFGpS method is highly modular, allowing for the use of distinct and diverse extracts to be mixed for the production of a variety of glycoproteins. “We are really excited about where this technology is headed,” Jewett said, “and think our new cell-free system has the potential to transform the way we make medicines.”
The work was supported by grants from the Defense Threat Reduction Agency and the National Science Foundation.
Original article published on July 12, 2018. Written by Tom Fleischman, Cornell Chronicle