Northwestern Engineering researchers have developed a new platform that can image single molecules in 3-D, allowing deeper probes into the inner workings of cells.
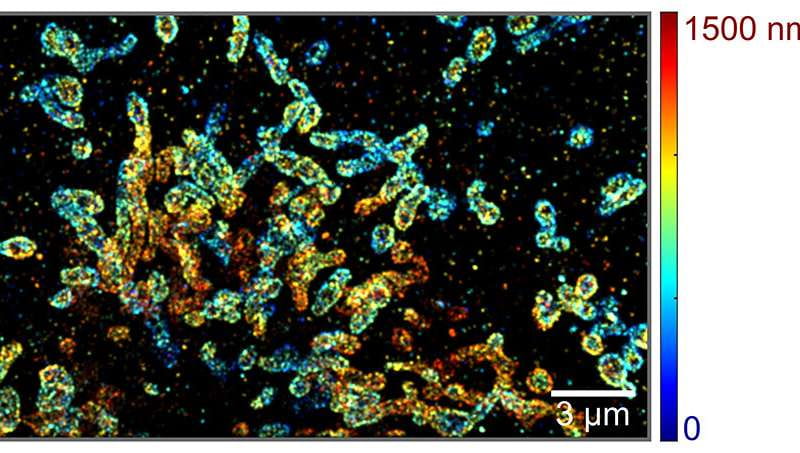
The platform uses spectroscopic single-molecule localization microscopy (sSMLM), a tool that can simultaneously capture the spatial informationof single moleculesand their spectroscopic signatures.
Researchers improved the tool by combining existing sSMLM with a two-mirror system, allowing it to image molecules in 3-D at much larger depths. This new tool could help molecular biologists understand complex processes inside cells.
“Our design is relatively easy to implement, and will allow us to study molecular interactions much better than before,” said Hao Zhang, professor of biomedical engineering and coauthor of the research. “Now we can not only see where molecules are, but also what they are.” Zhang developed the technology with Cheng Sun, associate professor of mechanical engineering.
The results were published May 21 in the journal Optica. Coauthors included Ki-HeeSong, a Ph.D. candidate, and Yang Zhan, a postdoctoral fellow, both of Northwestern’s Biomedical Engineering department.
Imaging in 3-D
In recent years, scientists and engineers have used sSMLM to better understand molecular interactions and cellular dynamics. The system provides information on the location of molecules and how those molecules interact with light, which tells scientists what type of molecule they see.
But the system only works in two dimensions, giving just a partial view of molecules and their interactions.
Zhang and Sun wanted to extend imaging to 3-D and originally developed a way to do this by adding an additional lens, but found that a pair of mirrors can achieve the same effect in a much more elegant way.
The mirrors work by introducing an optical path length difference in the system that improves the way the system uses photons. Unlike lenses, most mirrors do not attenuate light that is reflected, meaning more photons can be used for nanoscopic localization to create a sharper picture and extends imaging into 3-D-depth range.
With 3-D nanoscale imaging capability, researchers can see more interactions happening within the intracellular volume without being overshadowed by the surface. For example, Zhang, Sun, and their collaborators are using the system to study the intercellular distribution of molecules, looking at how RNA is transported and interacts with cell organelles before being translated into proteins.
“This system could have profound implications in molecular biology,” Zhang said.
Though previous molecular imaging systems used optical filters to detect different types of molecules based on their well-separated emission colors, the new system can detect minute differences in molecular emissions from every different molecule and analyze the spectrum to differentiate them.
“We can now color code every single molecule,” Sun said. “That is a key strength.”
Understanding nanoscale processes
Next, the researchers hope to continue to refine the technology, as well as use it in molecular biology studies.
They are working with collaborators to study the pore structure of the nucleus and how it is involved in stem cell differentiation, and are also studying the depolarization of the mitochondrial membrane, an event associated with many diseases, including vision loss in diabetic patients. They also hope that their technology helps others in the field.
“It is a very elegant design,” Sun said. “The system can be very easily implemented in other labs, and it has a beautiful performance.”
by Emily Ayshford, Northwestern University
The original story was published in Physics.org on May, 22, 2019.
Hao Zhang and Cheng Sun are both members of the Chemistry of Life Processes Institute.