News
Northwestern’s Chemistry of Life Processes Institute Receives a Grant Award from The Michael J. Fox Foundation
As is the case with many other diseases, a protein is thought to be a key driver of Parkinson’s disease (PD). Thanks to a two-year $700,000 grant from The Michael J. Fox Foundation for Parkinson’s Research (MJFF), researchers in Northwestern’s Chemistry of Life...

Student Spotlight: Ananya Basu
Ananya Basu, a third-year PhD student, is using innovative approaches to overcome previously intractable targets in the Zhang Lab. Appointed to the CLP Predoctoral Training Program, she is being co-mentored by Zhang (chemistry) and Horvath (molecular biosciences).
What is the focus of your research and how did you become interested in that area?
I work in the field of targeted therapeutics, specifically on the discovery of novel small-molecule protein degraders. I started as a high school student in a lab at Cincinnati Children’s Hospital where I was first exposed to high-throughout anticancer drug discovery in the context of pediatric leukemia, and this fostered an interest in therapeutics. I continued working in the Oncology department throughout my undergraduate years in preclinical drug development, as well as in synthetic chemistry labs in the Chemistry department at the University of Cincinnati studying organometallic catalysis and bioinorganic chemistry. These varied experiences led me to chemical biology for my graduate work.
Can you tell me about one of the highlights of your academic career?
Last July, I identified the first promising lead for targeted protein degradation of my graduate career. Data from follow-up with this lead was submitted as part of a disclosure to Northwestern’s INVO office, which was particularly exciting for me. It has also been very rewarding to mentor our Lambert fellow, Chi Li, and watch him not only come into his own in the lab but also build the foundation for what will be a very bright future as a scientist.
What is the most effective way to explain what you do to a nonscientist?
Many blockbuster drugs are well-designed inhibitors that block the activity enzymes that are involved in disease. Enzymes, because they carry out very precise chemical transformations, have defined structures that are well-suited for drug design. However, many proteins involved in the development or progression of cancer, for example, are not enzymes and lack structured regions where drugs can bind and block their function. Targeted protein degradation is an alternative strategy to access these disease-related proteins for therapy and is an exciting avenue for modern therapeutics. Degraders can bind disease-related proteins and cause them to be destroyed in cells, which blocks their activity. Identifying new degraders for disease-relevant proteins may help improve patient outcomes in the future.
You are part of the NIH Graduate Training Program with the Chemistry of Life Processes Institute in 2022-23. Could you tell us more about that program and how has it benefited you?
I am very fortunate to be a part of the Chemistry of Life Processes training program and have had many of my most positive graduate school experiences as part of the program. The program aims to train scientists to conduct rigorous research at the chemistry-biology interface, which is a perfect fit for my interests. I have had numerous opportunities to present my work through research forums to an interdisciplinary audience and network with other students and faculty, and these experiences have proven to be invaluable for me. The opportunity to develop orthogonal skills in a secondary mentor’s lab is also very unique. All of the staff and faculty involved in the training program are dedicated, supportive, and kind. Penelope, who helps coordinate many of our activities, is an absolute light in our lives and truly cares for us during and after our tenure in the program. Even as a nonscientist she has a deep understanding of the academic milieu and offers her continued and unwavering support.
What do you enjoy most about working with the Zhang Lab?
Joining a new lab is definitely a unique and formative experience. As an early member of a new lab, you have a role in setting the lab culture and I feel that our lab as a whole is a very tight-knit community. We take birthdays and holidays very seriously (as I write this our lab is in full Halloween spirit) and Xiaoyu has diligently promoted a Monday-morning bagel culture that I strongly approve of. Xiaoyu is a creative thinker with lofty aspirations for his research program and I am fortunate to work under his direction.
In 10 years, where do you hope to be in your career?
I think that the early-career period is the best time to take calculated risks, so I hope in ten years that I can say that some of those choices paid off. As a first-year, I took a course under Dr. Rick Morimoto and he advised me to always work on things with impact, and that really resonated with me. I think that high-risk, high-reward ideas are exciting and if I choose to continue in biotech post-graduation I think the start-up scene would be a good fit for me. If I hang up my lab coat for good, I might look towards a career in consulting or something more administrative. Most importantly, though, I see myself in ten years with an expansive walk-in closet and a vast array of sparkly possessions.
Aside from your work in chemistry, what hobbies do you enjoy?
The academic lifestyle is admittedly not very compatible with hobbies, but I love music, art, shopping, and good food. When I was younger, I played the violin quite often and enjoyed making cards for people in my life. Whenever I have the time and energy (unfortunately a rare co-occurrence) I like to explore nature or hop on the train to discover new places around Chicago.
This story was originally posted by Northwestern Chemistry 2023 Fall Newsletter.
Transfer learning paves the way for new disease treatments
Technological advances in gene sequencing and computing have led to an explosion in the availability of bioinformatic data and processing power, respectively, creating a ripe nexus for artificial intelligence (AI) to design strategies for controlling cell behavior.
In a new study, Northwestern University researchers have reaped fruit from this nexus by developing an AI-powered transfer learning approach that repurposes publicly available data to predict combinations of gene perturbations that can transform cell type or restore diseased cells to health.
The study was published today in the Proceedings of the National Academy of Sciences.
Since the completion of the human genome project 20 years ago, scientists have known that human DNA comprises more than 20,000 genes. However, it has remained a mystery as to how these genes work together to orchestrate the hundreds of different cell types in our body.
Surprisingly, essentially by guided trial-and-error, researchers have demonstrated that it is possible to “reprogram” cell type by manipulating only a handful of genes. The human genome project also facilitated advances in sequencing technologies, making it cheaper not only to read the genetic code, but also to measure gene expression, which quantifies the precursors of the proteins that carry out cell functions. This increase in affordability has led to the accumulation of a massive amount of publicly available bioinformatic data, raising the possibility of synthesizing these data to rationally design gene manipulations that can elicit desired cell behaviors.
The ability to control cell behavior, and thus transitions across cell types, can be applied to regrowing injured tissues or to transforming cancer cells back into normal cells.
Injured tissues resulting from strokes, arthritis and multiple sclerosis affect 2.9 million individuals each year in the United States, costing as much as $400 million per year. Meanwhile, cancers are responsible for around 10 million deaths annually worldwide with economic costs in the trillions of dollars. Because the current standard of care does not regenerate tissues and/or has limited efficacy, there is a critical need to develop more effective treatments that are broadly applicable, which in turn requires identification of molecular interventions that can be inferred from high-throughput data.
In the new study, the researchers train their AI to learn how gene expression gives rise to cell behavior using publicly available gene expression data. The predictive model generated by this learning process is transferred to specific cell reprogramming applications. In each application, the approach finds the combination of gene manipulations that is most likely to induce the desired cell type transition.
Unprecedented exploration of genome-wide dynamics
“Our work stands out from previous approaches to rationally design strategies to manipulate cell behavior,” said Thomas Wytock, lead author of the paper and member of the Center for Network Dynamics at Northwestern University. “These approaches mostly fall into two categories: one in which genes are organized into networks according to their interactions or common properties; and another in which the expression of genes from healthy and diseased cells are compared to single out the genes that show the largest differences.”
In the first category, there is a tradeoff between realism and scale. Some network models comprise many genes but only say whether a relationship is present or absent. Other models are quantitative and experimentally validated but necessarily involve a small number of genes and relationships. Northwestern’s new work retains the strengths of both types of models: it is inclusive of all genes in the cell and quantitative in representing their expressions. This is achieved by reducing the expression of nearly 20,000 individual genes to no more than 10 linear combinations of such genes, which are weighted averages referred to as eigengenes.
“Eigengenes basically show how genes operate in concert, making it possible to simplify the dynamics of a large dynamical network to just a few moving parts,” said Adilson Motter, the Charles E. and Emma H. Morrison Professor of Physics at the Weinberg College of Arts and Sciences, director of the Center for Network Dynamics at Northwestern University and the study’s senior author. “Each eigengene can be thought of as a generalized pathway that is approximately independent of the others. So, eigengenes pick up the relevant correlations and independences in the gene regulatory network.”
Approaches in the second category can find individual genes associated with a change in cell behavior but fail to specify how genes work together to enable this change. The new approach overcomes this challenge by recognizing that genes change their expressions in concert. The quantitative accounting of this property in terms of eigengenes makes it possible to additively combine their responses to different gene perturbations by suitably scaling them. The combined responses can then be input into the AI model to determine which perturbations elicit the desired cell behavior.
Averting combinatorial explosion
Equipped with this AI model, the researchers curated publicly available data to identify how gene expression changes when a single gene is perturbed by exogenously raising or lowering its expression. They then developed an algorithm to solve the inverse problem, which is to predict gene combinations that are most likely to induce a desired reprogramming transition, such as to cause diseased cells to behave as healthy cells. The approach that results from integrating the data and algorithm circumvents combinatorial explosion that would result from testing all combinations in order to identify the effective ones. This is significant because experiments can test only a limited number of cases, and the algorithm provides a way to identify the most promising cases to be tested.
“The approach shines in its ability to examine myriad combinations computationally,” said Wytock. “For example, the pairwise combinations of 200 perturbations yields 20,000 cases, triples yield over 1.3 million cases, and this number keeps growing exponentially. Because the algorithm employs optimization, the approach can compare predictions across a potentially infinite number of combinations through the magic of calculus.”
Another challenge circumvented by the approach is that the gene perturbations can combine in a non-additive manner. For example, consider the impact of gene perturbations on cellular growth rate and imagine perturbations halve the growth rate when applied in isolation. The effect of two such perturbations combine non-additively if they reduce growth to either significantly more or significantly less than half of a half (or one quarter). Even though there is a large body of research characterizing non-additive interactions between genes, the new approach is effective even without having to account for such deviations from additivity.
“This is a case in which the whole is well approximated by the sum of the parts,” Motter said. “This property of the interventions needed to induce transitions between cell types is counterintuitive because the cell types themselves emerge from collective interactions among genes.”
Because the approach addresses the main challenges to control cell behavior, it can be applied to many different biomedical conditions, including those that will benefit from future data.
A flexible model for forthcoming data
The fact that responses to gene perturbations combine additively facilitates generalization across cell types. For example, if a gene is disrupted in a skin cell, the resulting impact on expression would be largely the same in a liver cell.
Thus, the AI-powered approach can be thought of as a platform into which data pertaining to a specific disease in a specific patient may be inserted. The approach may be applied whenever curing the disease can be conceived as a reprogramming problem, as in the case of cancers, diabetes, and autoimmune diseases, which all result from cell dysfunction.
The versatility of the approach allows the gene expression in a single study to be rapidly contextualized across all available data in the National Center for Biotechnology Information’s Sequencing Read Archive, which is the largest publicly available repository for gene expression data. This archive has grown 100-fold from 10 terabytes to 1,000 terabytes between 2012 and 2022 and continues to grow exponentially as sequencing costs decrease. This work provides a critical tool for translating this wealth of data into specific predictions of how genes work together to control the behavior of normal and diseased cells.
The study, “Cell reprogramming design by transfer learning of functional transcriptional networks,” was supported by the Army Research Office, National Institutes of Health through the Chicago Region Physical Sciences Oncology Center, led by the Chemistry of Life Processes Institute and Robert H. Lurie Comprehensive Cancer Center of Northwestern University, National Science Foundation and Northwestern University’s Malnati Brain Tumor Institute.
Northwestern Now|March 4, 2024 | By Ellie Mejía

Neil Kelleher to Headline and Receive Distinguished Contribution Award at US HUPO 2024
Neil Kelleher, Walter and Mary Elizabeth Glass Professor of Chemistry, Molecular Biosciences, and Medicine at Northwestern University, will present his groundbreaking work and insights at US HUPO 2024 in Portland, Oregon next week as the 2024 recipient of the Donald F. Hunt Distinguished Contribution in Proteomics Award.
“Dr. Kelleher epitomizes a focused achievement within the realm of proteomics,” stated HUPO leadership in their press announcement. “This award recognizes his unparalleled contributions to targeted applications in top-down proteomics, natural products discovery, and chromatin biology, and celebrates Kelleher’s impact on proteomics research.”
Kelleher, the Director of Northwestern’s Chemistry of Life Processes Institute and its 50-person Proteomics Center of Excellence, will speak at US HUPO 2024 on Monday, March 11, 2024, from 8:30 AM to 9:05 AM PT. In addition to celebrating the legacy of Professor Donald F. Hunt, whose approach to science, technology and mentorship has had an enormous impact on the field, Kelleher will explain his pioneering approach to detecting and characterizing human proteins in his plenary talk, ‘Harnessing Mass Spectrometry to Help Domesticate the Human Proteome.’
For over twenty years, the Kelleher Group has invented new methods to discover the exact forms of protein molecules in human cells. The world has come to call these “proteoforms.” Kelleher uses “Top-Down” Proteomics to discover, characterize and assign function to them with increasing efficiency. The “domestication” of the human proteome via precise compositional mapping will improve the efficiency of basic and clinical research and therefore enhance diverse goals for the 21st Century, including designer organs, personalized medicine, and early detection of human disease. A recent article in Science (2022, 375: 411-418) typifies the promise and crescendo of activity in this area of proteomics, advanced consistently by Kelleher over the past 25 years.
A non-profit association, US HUPO (US Human Proteome Organization) engages in scientific and educational activities to encourage the use of proteomics technologies and to disseminate knowledge about the human proteome and that of model organisms. Learn more about US HUPO at www.ushupo.org.
by Lisa La Vallee

At the Human Longevity Lab, Studying methods to slow or reverse aging
The Potocsnak Longevity Institute at Northwestern University Feinberg School of Medicine has launched the Human Longevity Laboratory, a longitudinal, cross-sectional study that will investigate the relationship between chronological age and biological age across different organ systems and validate interventions that may reverse or slow down the processes of aging.

CLP Seminar
Identifying Latent Functional Dynamics in Proteins Using Bio-NMR
Jordan H Chill
Associate Professor
Bar Ilan University
I lead the high-resolution biomolecular NMR research group at Bar Ilan University, Israel, following a post-doctoral fellowship at the NIH under direction of Dr. Adriaan Bax. I am a structural biologist investigating protein structure, dynamics and function in health and disease, with current projects including inhibitory complexes of peptide toxins and potassium channels, structural biology of Wiskott-Aldrich syndrome, and SH3-targeting drug-design. Bio-NMR with its unique abilities is our primary tool, and we combine it with biophysical and computational methods to realize our research goals.
When: Friday, Feb. 9, 2024, 12:00 – 1:00 pm
Where: Pancoe Auditorium

Neuroscientist William Klein’s purpose and perseverance continues inspiring a new wave of efforts to combat Alzheimer’s disease
Battling Alzheimer’s disease has become William Klein’s life work – even if he never saw it coming.
A progressive disease impacting memory, thinking, behavior, and other mental functions, Alzheimer’s affects an estimated 6.7 million Americans, according to the Alzheimer’s Association, and that number is expected to soar to 13 million by 2050. The disease stifles quality of life for the afflicted, places a heart-wrenching, time-consuming burden on caregivers, and strains the nation’s healthcare system to the annual tune of $345 billion.
Over the last two decades, Klein, a professor in the Department of Neurobiology at Northwestern University’s Weinberg College of Arts and Sciences, has taken aim at Alzheimer’s disease with unrelenting purpose and an enterprising spirit. Blending his own scientific energy with earnest collaborators from Northwestern, the U.S., and abroad, Klein has fundamentally altered the course of Alzheimer’s research, diagnosis, and treatment, including the development of a novel antibody for treating the disease based on research initiated in his Hogan Hall laboratory.
After working under a pair of Nobel Prize-winning scientists – Paul Boyer at UCLA and Marshall Nirenberg at the National Institutes of Health – Klein arrived at Northwestern in 1976. Klein’s research focused on synapses – the places where neurons connect and communicate with each other. “I was enthralled by synapses and their role in all the amazing things our brains do, including learning and memory. I wanted to know how they were put together, how they adjusted, and how they ended up looking the way they do.” Yet for as fascinating as that research was, Klein wanted to apply his research to real-world problems. “It seemed to me it was feasible to take what I’d learned as a basic scientist and extend that to things that could make a difference in people’s lives.”
Klein remembers learning about the first Alzheimer’s patient, a German woman named Auguste Deter. When neuropathologist Dr. Alois Alzheimer asked Deter to pick up a pen and write her name, she couldn’t and famously said, “I have lost myself.” The story stuck with Klein, who believed he could tie his research to Alzheimer’s. “You know who you are thanks to the memories you have. When those start to dissolve, you’re no longer yourself. I thought I could do something to help.”
Klein’s first move into work with Alzheimer’s disease began in the early 1990s thanks to – of all things – a coffee maker. After Northwestern received a large grant from the state of Illinois to facilitate collaboration with local pharmaceutical companies, the University hired Catherine Propst, formerly of Abbott Laboratories, to lead the effort. To welcome Propst to Northwestern, Klein delivered a new coffee maker to her office, where the two discussed his research. A month later, Propst connected Klein, who was in the early stages of drawing a correlation between synapses and Alzheimer’s disease, to an Abbott Labs’ medicinal chemist named Grant Krafft. Klein and Krafft would become long-time collaborators on Alzheimer’s research, particularly after Krafft joined the pharmacology faculty at Northwestern’s Feinberg School of Medicine. “Who knew a ubiquitous biochemical instrument would help kick this off?”
Klein’s two decade-long run studying synapses and neurotransmitter receptors translated remarkably well to Alzheimer’s disease. “It turns out that the best pathological correlate of dementia and Alzheimer’s disease is loss of synapses.”
Alongside Krafft, Klein also teamed up with Caleb “Tuck” Finch, an eminent molecular biologist based at the University of Southern California. Through various experiments, the scientific trio posited that amyloid beta oligomers, not amyloid plaques as commonly thought, were the actual toxins responsible for Alzheimer’s disease. Those findings launched a company, Acumen, in 1996 and the group published its first paper in the Proceedings of the National Academy of Sciences (PNAS) in 1998. To date, that single paper has been cited more than 4,500 times.
Initially, however, the group’s boundary-breaking work struggled to gain a foothold in the scientific community. Klein recalls standing at a conference and overhearing a group of “very productive fellows” from a prestigious American university questioning the validity of his team’s results. Even after the researchers published a second PNAS paper in 2003 demonstrating amyloid beta oligomers existed in the Alzheimer’s brain, but not the control brain, skepticism remained. “For a long time, we had to be the salmon swimming upstream, and it was very, very difficult to get people to buy into the notion that the actual culprit [of Alzheimer’s disease] was amyloid beta oligomers.”
As Klein and his team shopped the potential for a receptor target to pharmaceutical companies, they encountered multiple dead ends. Promising deals collapsed and questions persisted about drug discovery’s ability to tackle Alzheimer’s. Disappointment began mounting and progress seemed slow, if not wholly elusive. “That leads to the question from my wife: ‘What’s taking so long?’”
Still, Klein and his collaborators continued pushing. They earned funding to power additional experiments; presented their findings at conferences; nurtured collaborations with investigators near and far; and entered an exclusive license and research collaboration with Merck & Co. They also created the first antibodies to amyloid beta oligomers and willingly shared those with other scientists to fuel additional experiments. Klein, in particular, stayed committed to the mission. “You just can’t roll over. The data was on my side, and I wasn’t going to give up.”
By the early 2010s, the group’s science gained wider approval – and the momentum has only accelerated over subsequent years. Today, laboratories and pharmaceutical companies around the world are studying amyloid beta oligomers and developing therapeutics to target the toxic proteins. Klein’s Northwestern-based lab, meanwhile, has collected a combined 170 papers and patents on the topic. “The dominoes are falling as people acknowledge amyloid beta oligomers look to be an authentic target for Alzheimer’s therapeutics.” In fact, over 5,000 papers have been published on amyloid beta oligomers and publications from Klein’s lab have been cited more than 35,000 times.
Acumen is among the foremost drug discovery companies attacking amyloid beta oligomers and its most promising therapeutic, ACU193, derives from an antibody program initiated in Klein’s research lab decades ago. In 2021, ACU193 entered a Phase 1 clinical trial and earned favorable results. Last November, BioTech Breakthrough, an independent organization recognizing standout life sciences and biotech companies, awarded Acumen and ACU193 its prestigious “Monoclonal Antibody Solution of the Year” as part of its BioTech Breakthrough Awards program. ACU193 is now preparing for a phase 2/3 trial with the U.S. Food and Drug Administration. “We’re finally on the fast track.”
Klein, who also has an appointment in the Department of Neurology at Feinberg, praises Northwestern’s intellectual horsepower, collaborative energy, and entrepreneurial ecosystem. He credits collaborations with some of Northwestern’s top scientists for propelling his team’s progress and spurring a deeper understanding of Alzheimer’s and how to combat the disease. Over the years, he’s partnered with renowned colleagues from Weinberg’s Department of Chemistry, such as Chad Mirkin, Neil Kelleher, Rick Silverman, Thomas Meade, and Milan Mrksich as well as researchers from Feinberg and the McCormick School of Engineering like Vinayak Dravid. Klein has also leveraged emerging resources at Northwestern, from imaging technologies to the Keck Biophysics Facility, to drive investigations and leaned on a capable team led by lab manager Kirsten Viola, who has worked on the Alzheimer’s project since its inception. “We have the intellectual camaraderie and the resources in terms of instrumentation at Northwestern to really help scientists like me make a difference.”
Five decades into his scientific career and nearly 30 years into probing the mysteries of Alzheimer’s to generate a molecular basis for the cause, diagnosis, and treatment of the disease, Klein remains as passionate and determined as ever. “It’s the chasing after it that inspires and the idea that I can really make a difference that continues to motivate me.”
Click here to view the original story.
Inside the chase after those elusive proteoforms
Human cells contain crowds of protein variants, but, especially in a time of funding challenges, chasing these proteoforms takes dogged persistence.
Biology delivers a massive number of puzzles to proteoform hunters1,2. Proteoforms are the droves of protein variants that one and the same gene can give rise to. This variant explosion takes place after DNA is transcribed to mRNA and a protein is synthesized. No single method currently lets researchers hunt these proteoforms reliably and at scale. Sparks fly in discussions about which of the existing approaches — ‘bottom-up’ or ‘top-down’ proteomics, shades thereof, or other, newer technologies — is most promising. Some rifts seem to be closing as scientists keep up their proteoform hunt in a tense funding environment.

When Instinct and Passion Collide
A longstanding member of Chemistry of Life Processes Institute’s Executive Advisory Board, Steven Deitcher, MD, Founder, CEO, and Chairman of Bespoke Biotherapeutics, has helped advance the Institute’s mission to conquer disease through protein-informed precision medicine. An accomplished academic and business leader, Deitcher earned his bachelor’s degree in medical sciences (a six-year program offered at the time), and his MD from Northwestern University Medical School. His work spans medicine and private and public company C-suite biomedical product development and commercialization. Among his many contributions to the Institute, Deitcher and his family foundation generously support CLP’s Convergence Workshops which bring together world-class teams of chemists, life scientists and engineers with clinicians in Northwestern’s Feinberg School of Medicine to tackle major clinical challenges with protein-informed approaches.
Where did your love for science begin?
I grew up in Lynnfield, Massachusetts, and always loved science and mathematics. I would describe myself as a very curious and mechanically inclined individual. Curious to the point of getting in trouble with family, teachers, and clergy, because I liked asking the questions “why” and “how”. As a child, I was fascinated by the human body and biology. At a very young age, relatives would buy me books. I think I got my first Grey’s Anatomy book when I was around five and my first Human Body book when I was six years old. I still have these books from my childhood. Interestingly, I can look at the chapters that most fascinated me back then because they’re either dogeared or highlighted as a five or 6-year-old. Those are the exact areas of medicine I ended up focusing my investigative, academic, and industry careers.
I was particularly interested in the chapters on the lymphatic system and the circulatory system, and I thought that white blood cells were the coolest things in the world. Fast forward 50+ years and I enthusiastically spend my days engineering white blood cells to combat cancer and treat hemophilia A.
Why did you choose to attend Northwestern?
When I was looking at colleges and universities, I came across the Honors Program in Medical Education (HPME) at Northwestern. There were several combined or accelerated programs at other schools, but when I started digging, I realized that Northwestern’s six-year medical program was one of two original accelerated programs and oriented toward clinical medicine. There was also a published ranking of honors programs in medical education and Northwestern was ranked number one. So, I applied to it. I remember during my interview being asked if I had applied to all the different six-year medical programs and I said, “No, I picked the one I want to go to. I want to come here.” I think that helped me get in.
The day I interviewed at the medical school; the entire city of Chicago was shut down. The windchills were as low as -70 to -80 degrees and the streets were coated with ice. I remember flying in from Boston with my father and he looked at me and said, “Look at the weather! Are you sure this is where you want to come?”
How was your experience at Northwestern?
I loved it. I enjoyed the academic freedom, the ability to take senior-level courses even as a freshman, and the exposure to fellow students from all over the US who were super bright and ambitious. I loved Evanston and the feeling of Chicago and the medical school. While on the Evanston campus, I would go down to Chicago as frequently as I could. Sometimes when I come to CLP meetings, I’ll stay in Chicago and take the “L” up to Evanston.
While I was in my second year of medical school, my mother passed away. This stimulated me to pursue my own path in medicine. I had things I wanted to do. My interest was using immune cells and manipulating them to fight breast cancer which is what my mother died from. So, I took some time off from school, received some grant money, and secured lab space and mentorship at Rush. This initial exposure to hematopoietic stem cell research evolved into an extracurricular activity that spanned my entire last two years of medical school. I used to go from medical school at Northwestern in the Streeterville neighborhood to spending the whole night in the lab in the Illinois Medical District, running experiments, then coming back to my apartment in River North to change before heading back to school. Everything that I was doing simply captivated and motivated me.
[During this time Deitcher also took night classes at Northwestern’s Kellogg School of Management.]
I have vivid memories by daytime sitting in the lecture hall in medical school and learning about the tests you should run to monitor someone with lupus and at night there’d be a lecture by someone at the business school on when you should stop doing tests in people with lupus because you’ve reached the point of diminishing financial returns. At this point, I knew that I wanted to save lives more than save money.
What did you do after Northwestern?
I applied for and completed my internship and residency at Barnes Hospital at Washington University in St. Louis which was known as one of the most intensive internal medicine residencies in the country. That appealed to me. During residency, I invented my first product, a bedside diagnostic test to rule out gram-negative bacteremia which got approved and published my first original research paper. I actually first met the CLP EAB Chair, Dr. Andy Chan, during my internship. After three years, I left St. Louis and went back to Boston where I did clinical fellowships in hematology and medical oncology (at Tufts University School of Medicine and New England Medical Center Hospitals) and then completed postdoctoral research training in coagulation protein chemistry.
At age 30, I left and took my first full-time faculty position at the University of Tennessee Health Science Center and St. Jude Children’s Research Hospital. My ambition and outspokenness rapidly landed me the additional positions of vice chairman of the Department of Medicine and assistant dean for clinical education. I spent four full years in Memphis before relocating to a dream position at the Cleveland Clinic. I spent the last six-plus years of my academic career as the head of the Section of Hematology and the director of vascular medicine research. These were transformative years.
What made you leap from Medicine to Biotech?
I had the opportunity to leave and join the leadership team of a biotech company in California that had licensed some technology and a novel protein therapeutic from a larger company that I had worked on while I was at the Cleveland Clinic. I remember getting the offer. I was 39 and thriving in a highly desirable position at a great institution. One of my faculty came into my office in December of 2003, shook his head, and said, ‘You’re so lucky. You can just cruise for the next 25 years and do the same thing you’re doing now.” And it felt like lightning hit me. I realized that I didn’t want to cruise, that to make a giant impact and do something significant in medicine, I had to get a new drug approved or discover something that could be applied to a population of patients, not just seeing patients one at a time. It was exciting and new and forced me to think differently and learn a lot. I’m now coming up on my 20th anniversary of joining the biotechnology industry in California. I wake up every day excited to dive into my work and continue to steer clear of boredom.
Why did you join CLP’s Executive Advisory Board?
CLP’s leadership, members, and executive board are very congenial, energetic, accomplished, and intelligent groups of people. As a protein chemist, a physician, an entrepreneur, and a person who’s done a lot in the past and never liked to be viewed as one-dimensional, CLP was very attractive because of its multidisciplinary research approach. I believe in the mission where diversity of thought, perspective, and approach is conducive to better healthcare solutions.
CLP’s strong relationship with Northwestern’s Feinberg School of Medicine is appealing to me because it allows me to be involved in something that makes me feel like I’m part of a ‘One Northwestern’ effort, not just an Evanston campus effort or a Chicago campus effort. I try to be an active part of the valuable and necessary “bridge” between campuses.
CLP takes high-risk, high-reward approaches to life-threatening conditions. You need investigators representing a broad array of disciplines to accelerate the discovery and development of such advances in the diagnosis and treatment of disease. We also need people who are taking a much more scientifically entrepreneurial—a big paradigm shift approach to things. If it doesn’t work, okay. Doesn’t work. If it does work, we’re not making a little incremental improvement, we’re significantly moving the needle. I find that exciting. A lot of the discussion we have at CLP is about how to make a big difference, how to generate solutions, and how to look at the questions a little differently than others may look at them. I am honored to be part of a group that is so passionate about science and focused on change.
What do you think has been the key to your success?

Deitcher, a father of four who lives in the San Francisco Bay area with his wife and youngest children, is pictured here with his equestrian daughter, Natalie, and her horse, Snap.
I always believed in following my own instincts. The work I did between 1986 and 1988 serves as the foundation of what my company [Bespoke Biotherapeutics] does today. There was a big gap in between because technology limitations prevented us from genomically engineering immune cells in the 1980s, 1990s and 2000s. I was involved in the very early attempts to harness cell therapy—basically, bone marrow transplantation—for women with metastatic breast cancer. It didn’t provide much benefit but certainly caused significant toxicity.
The lab team would say, “We need to figure out how to reprogram the immune cells.” Technology was so far from being able to allow us to do that back then, but now we can. And that’s what I’m doing. Bespoke’s lead oncology product candidate is genetically engineered B-cells targeting breast cancer.
Sometimes early research arcs resurface later in your life, and they make you smile. I wake up every morning wanting to succeed at the work that I’m involved in. It keeps me stimulated and interested. I won’t say that money isn’t important, but it isn’t the most important thing. To me, not being bored is the most important thing. I don’t like wasting time and I like to be invigorated.
by Lisa La Vallee
CLP Lambert Fellows Lean into Drug Discovery
For 13 years, the Chemistry of Life Processes Institute has provided financial support for promising undergraduate Chemistry majors to conduct research for two consecutive years through the Institute’s Lambert Fellowship Program. This year, three new fellows, Monica Jones, Young Chi Li, and Daniel Yang joined the current cohort that includes 4th-year students JoJo Holm, and Anthony Tam.
In addition to a stipend that covers their living expenses, Lambert Fellows also receive laboratory funding that covers research materials and supplies as well as travel and academic conference fees. The Fellowship also includes a mentoring plan that allows students to develop their skills with CLP faculty members and postdoctoral and graduate students who share the same interests.
Recently, CLP spoke with awardees about their research projects, experience in the fellowship program and plans for the future.

MONICA JONES
Hometown: Austin, TX
Year: 2nd
Major: Chemistry
CLP Mentor: Regan Thomson (Chemistry)
What is the focus of your Lambert Fellowship Research Project?
In the past five years or so, there has been a wave within the drug synthesis community to recreate a lot of drugs that we already produce with changes, such as exchanging a hydrogen atom for a heavier deuterium atom. Many promising studies have shown that drugs with a change in that atom would stay longer in the human body and would allow people to take fewer doses of medication. This provides another avenue for developing extended-release 24-hour medications.
We think that if you exchange some of the components that just have hydrogen in them for deuterated components, then you would be able to add deuterium at a certain location on a molecule that you’re using for drug synthesis. The mechanisms I used during the summer had about a 40% yield which wasn’t great. It wasn’t what we were looking for. However, I did find a mechanism that uses a catalyst that we thought could have potential as well as other projects with biocomponents that we would like to investigate further.
Why did you apply for the Lambert Fellowship?
Originally, I applied for the CLP Summer Research grant because I wanted to do research last summer, but I didn’t have the money. However, after discussing the project with my professor, Regan Thomas, we realized that it would take longer than a summer, so he encouraged me to apply for the Lambert Fellowship instead. What it has meant to me is stability within this lab and within this project. I can keep working for a long time. My professor also really appreciated that extra funding for supplies.
Do you know yet what you want to do after completing your education?
I want to get a PhD at some point. Initially, I thought I wanted to go directly into a graduate program, but I’m not sure if that’s the best idea anymore because they don’t get paid very much and it would be good to work and build some industry connections. Eventually, I’d like to work in the pharmaceuticals industry, or possibly for a food-related or cosmetic company. But I want to stay in organic chemistry. Since I also work in the [Franz] Geiger lab, I have some interest in spectroscopy as well.

YOUNG CHI LI
Hometown: Hong Kong
Year: 3rd
Major: Chemistry and Integrated Science
CLP Mentor: Xiaoyu Zhang, PhD (Chemistry)
What is the focus of your Lambert Fellowship Research Project?
I screen for drugs that can degrade androgen [sex hormone family to which testosterone belongs] receptor variants in prostate cancer. Improper hormone signaling is one of the factors contributing to the development of prostate cancer. The first line of defense in treating prostate cancer is chemical or physical castration to lower the secretion of androgen. However, this isn’t a permanent solution. A second line of defense is androgen antagonist drugs, but they work only for a short time. Eventually, the cell creates variants of androgen receptors and the hormone signaling continues. My project is to find drugs to target these receptors.
Why did you apply for the Lambert Fellowship?
I worked in Professor Zhang’s lab in the summer of my freshman year and I really enjoyed working with him, so we decided to make a long-term plan to apply for the Lambert Fellowship because it offers two years of support. He also thought that the fellowship could give me some experience in research, whether going into industry or academia. I also wanted to meet more people through the Fellowship and connect with alumni, so I decided to apply.
Do you know yet what you want to do after completing your education?
I am probably going to pursue computer science or finance: something related to math or tech.

DANIEL YANG
Hometown: Webster, NY (born in Shenzhen, China)
Year: 3rd
Major: Chemistry, Biology and Integrated Science
CLP Mentor: Karl Scheidt (Chemistry)
What is the focus of your Lambert Fellowship Research Project?
My research project targets an enzyme that plays a key role in pancreatic cancer metastasis. We are trying to modify a chemical inhibitor to shut down the pathway for metastasis. By binding to an upstream signaling kinase, we hope to shut down that enzyme, blocking the signal that calls for cell proliferation from reaching the end of the pathway and causing uncontrolled cell division. My project is focused on modifying the inhibitor’s chemical structure to make it more potent and selective for that specific kinase.
Why did you apply for the Lambert Fellowship?
Through my junior year, I worked with Professor Scheidt and was able to explore a variety of organic chemistry projects, which ultimately led me to medicinal chemistry. There, I realized that the project required long-term commitment and thus applied to the Lambert Fellowship. Through it, I have gained an understanding of what it is like to work in an academic lab. The Lambert has also given me the opportunity to talk to other scholars that are my age through the different conferences that CLP plans.
Do you know yet what you want to do after completing your education?
I’d like to go into medicine. I think this project has helped me build a good foundation for medical studies and solidified a connection between synthetic chemicals and biological pathways.

Second year fellows, JoJo Holm and Anthony Tam, plan to go back to school after graduating from Northwestern. JoJo intends to apply to medical school, while Anthony will pursue a PhD in chemistry.
The Lambert Fellowship was founded in 2010, and endowed in 2016, by CLP Executive Advisory Board Chairman and Senior Vice President of Research Biology at Genentech, Andrew Chan, MD, PhD (Weinberg). The program is named in honor of Dr. Joseph B. Lambert, the Clare Hamilton Hall Professor of Chemistry. Lambert was Dr. Chan’s advisor in the late 1970’s at Northwestern and helped him succeed as a student; Lambert had a profound impact on Chan’s educational experience.
by Lisa La Vallee

CLP Trainee IDP Workshop
Sheila Judge, PhD
CLP Senior Director for Research, Education and Administration
Sheila Judge will lead a hands-on, interactive workshop, using the MyIDP website offered by Science Careers to create an individual development plan. Trainees will take an online skills assessment, along with an interest assessment, which will lead to creating professional goals to work on in cooperation with their mentors.
When: Monday, Dec. 4, 2023, 4:00 – 5:30 pm
Where: Silverman #1531
Pioneering Automated Proteoform Imaging of Ovarian Cancer Tissue
Investigators led by Neil Kelleher, PhD, professor of Medicine in the Division of Hematology and Oncology and of Biochemistry and Molecular Genetics, have developed an automated technique for imaging and identifying proteoforms in ovarian cancer tissue, according to results published in Nature Communications.
The technique offers the greatest speed and accuracy currently available for the high-resolution, high-throughput imaging of proteoforms—all the modified versions of proteins in a tissue sample—and has multiple potential applications in cancer diagnostics, Kelleher said.

Neil Kelleher, PhD, professor of Medicine in the Division of Hematology and Oncology and of Biochemistry and Molecular Genetics, and director of the Chemistry of Life Processes Institute
Neil Kelleher, PhD, professor of Medicine in the Division of Hematology and Oncology and of Biochemistry and Molecular Genetics, and director of the Chemistry of Life Processes Institute, was senior author of the study published in Nature Communications.
Several techniques are currently used to image proteins in human tissue, but very few are capable of imaging proteoforms. Those that can sample proteoforms directly from tissue do so by ionizing them for mass spectrometry. Other techniques prevent scientists from understanding where the proteoforms exist within a tissue and do not identify all existing proteoforms in a sample at once.
To better understand the spatial position of proteoforms within a given tissue, Kelleher’s team developed proteoform imaging mass spectrometry (PiMS), which was detailed in a 2022 study published in Science Advances. The technique works by sampling proteoforms from the tissue with nanodroplets — “weighing” the extracted proteoforms to identify those up to a certain size and then using this data to construct proteoform images of the scanned tissue.
In the current study, Kelleher and his collaborators built upon this technique to create AutoPiMS, which utilizes a computational engine to automatically identify and characterize proteoforms within a thin section of cancer tissue.
AutoPiMS was able to identify and characterize more than 300 proteoforms within a human ovarian cancer tissue sample, and mapped where specific cancer-associated proteins existed within the sample at a speed of one minute per proteoform. AutoPiMS was also able to identify cancer tissues versus non-cancerous tissues from the same patient.
“This technique is really important for cancer tissue imaging and cancer diagnostics,” said Kelleher, who also directs the Proteomics Center of Excellence and the Chemistry of Life Processes Institute. “We have shown in this paper that we can locate not just proteins, but their myriad proteoforms, the ultra-specific kind of measurement in my field of proteomics.”
The technique will be made available to other Northwestern proteomics investigators, Kelleher said, and he hopes AutoPiMS will accelerate discoveries in the field.
Moving forward, Kelleher and his collaborators will adapt the technique for use in single-cell proteomics, he said.
“If we can have information on single-cell proteoforms, we would have the most precise information about proteins in space, time and composition. That’s the ultimate technology that would make protein analysis so much more precise,” Kelleher said. “Precision medicine requires precision proteomics. The more precise we can make it, the more we can advance drug development, lower side effects and improve diagnostics, all of which are aligned with the new mission of the CLP Institute at Northwestern.”
Kelleher is also a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
The study was supported by National Institutes of Health grants P41 GM108569, P30 DA018310, P30 CA060553 and F31 AG069456, as well as funding from the U.S. Department of Defense Uniformed Services University of the Health Sciences.
by Olivia Dimmer
This story was published by Feinberg News on Nov 9, 2023.

CLP Welcomes Outstanding 2023-24 NIH Predoctoral Training Program Cohort
This fall, the Chemistry of Life Processes Institute welcomed a new cohort of Northwestern graduate students to its NIH Predoctoral Training Program. Alondra Sanchez, Ana Leal, Assa Magassa, Jun dos Remedios, Miguel Campos, and Yara Jabbour Al Maalouf joined second-year trainees Amy Tang, Ananya Basu, and Daniel de Castro Assumpcao, in the prestigious program to conduct immersive research experiences that span the interface between biology, chemical biology, engineering, and organic, inorganic and physical chemistry. CLP trainees are distinguished by their strong commitment to interdisciplinary research, research experience and productivity, and their ability to overcome obstacles to pursue their career goals.
Trainees select two mentors from a roster of 54 highly interdisciplinary Northwestern faculty whose research programs fall within the following areas: development of molecular therapeutics and diagnostics; study of molecular mechanisms and targets; molecular and systems modeling; and engineering of biological systems. They are required to spend at least 10-weeks in the lab of their secondary mentor to learn new methods and approaches.
“The CLP Predoctoral Training Program provides students with a unique opportunity to bridge the gap between theoretical knowledge and hands-on experience in the field of chemical biology. It also equips students with a strong support system,” says CLP member Xiaoyu Zhang (Chemistry), a preceptor of the program.
CLP trainees must complete two courses that provide the intellectual foundation and hands-on experience needed to be successful in working across the chemistry-biology interface. Participation in monthly research forums and communications workshops helps students build confidence and hone their presentation skills. As part of the program, trainees also invite and host prestigious scientists from across academia and industry to give lectures at Northwestern.
“It’s a delight to see students pursuing ambitious, interdisciplinary projects through CLP! I love seeing students grow out of their comfort zones through collaborations with other fields,” says CLP faculty member and program preceptor Gabriel Rocklin (Pharmacology).
This year’s new graduate trainees are:

Alondra Sanchez
Graduate Program: Chemistry
Preceptors: Xiaoyu Zhang (Chemistry) and Deyu Fang (Pathology)
 Ana Leal
Ana Leal
Graduate Program: Chemistry
Preceptors: Chad Mirkin (Chemistry) and Bin Zhang (Cancer Immunology)
 Assa Magassa
Assa Magassa
Graduate Program: Chemistry
Preceptors: Xiaoyu Zhang (Chemistry) and Gabriel Rocklin (Pharmacology)
Jun dos Remedios
Graduate Program: Chemistry
Preceptors: Richard Silverman (Chemistry) and Richard Morimoto (Molecular Biosciences)
 Miguel Campos
Miguel Campos
Graduate Program: Chemistry
Preceptors: Karl Scheidt (Chemistry) and Xiaoyu Zhang (Chemistry)
 Yara Jabbour Al Maalou
Yara Jabbour Al Maalou
Graduate Program: Chemical Engineering
Preceptors: Keith Tyo (Chemical and Biological Engineering) and Gabriel Rocklin (Pharmacology)
The 2nd-year graduate trainees are:

Amy Tang
Graduate Program: Driskill Graduate Program in Life Sciences
Preceptors: Deyu Fang (Pathology) and Xiaoyu Zhang (Chemistry)

Ananya Basu
Graduate Program: Chemistry
Preceptors: Xiaoyu Zhang (Chemistry) and Curt Horvath (Molecular Biosciences)
 Daniel de Castro Assumpcao
Daniel de Castro Assumpcao
Graduate Program: Chemical Engineering
Preceptors: Danielle Tullman-Ercek (Chemical and Biological Engineering) and Neal Devaraj (Biochemistry and Biophysics)
“I believe the program plays a vital role in shaping the next generation of chemical biologists,” says Zhang. “I am proud to be part of this journey and am excited to witness the program’s continued success.”
by Lisa La Vallee

CLP Summer Scholars Learn Research Fundamentals
Since its inception in 2009, the Chemistry of Life Processes Institute (CLP) has nurtured the research interests of dozens of undergraduates through its Summer Scholars Program. Program participants not only benefit from outstanding CLP faculty mentors; they receive training in the latest scientific methods, state-of-the-art instrumentation, and laboratory best practices. The Program supported three students this year: Gabriela Cosenza, Mychaela Mathews, and Adam Suh. They received stipends and research support through the generosity of donors to the Institute. The Scholars were selected through a competitive application process that weighed students’ research experience, aptitude, interest, endorsement by their CLP faculty member, and the interdisciplinary nature of their research.
“We love offering undergrads the opportunity to spend the summer on campus working full-time in CLP faculty labs. It gives them a chance to see how working in a lab professionally would be once they graduate. Plus, interacting with other students who plan to pursue advanced science degrees helps them build a network for the future,” Penelope Johnson, CLP Senior Project Coordinator, Education and Outreach. “It’s inspiring that these students choose to stay on campus to further their education. We are very proud of their work ethic and their dedication to learning. I love our students!”
CLP’s Summer Scholars Program serves as a launchpad for students who wish to pursue careers in chemistry, engineering, life sciences, and medicine—82% of graduated CLP scholars have pursued or received advanced degrees (MS, PhDs, and MDs). One of the hallmarks of the program is its commitment to supporting underrepresented students. The program provides students with a summer stipend and funds for lab supplies to devote 10 full weeks of hands-on interdisciplinary biomedical research in the laboratory of one of the Institute’s 75 renowned faculty.
Seminars and workshops sharpen students’ science communications skills and prepare them for presenting the outcomes of their research at the annual CLP Undergraduate Research Symposium. This year, the students also met with representatives from Northwestern’s career services, students in the MD/PhD program at Feinberg, and current graduate students so they could learn more about possible career paths once they graduate.
Another benefit for students in the program is the opportunity to publish their research findings. CLP Summer Scholars Program have contributed to 31 research papers as undergraduates and 95 publications in post-graduate positions and are well-prepared for a successful entrée into science careers.
This year’s CLP Summer Scholars were:

Gabriela Cosenza
Gabriela Cosenza
Hometown: Honduras, Central America
Year: 2nd Year
Major: Chemical Engineering
Minor: Biotechnology and Biochemical Engineering
CLP Mentor: Joshua Leonard (Chemical and Biological Engineering)
What was the focus of your summer research project?
The Leonard lab focuses a lot on working with synthetic biology and cell therapies. Synthetic receptors are very useful in improving cell therapies and immunotherapies for diseases such as cancer. A common problem with synthetic receptors is that they can be single signaling—meaning once they fulfill their purpose, they don’t degrade naturally, or at least they don’t do it fast enough, so they can act as competitive inhibitors to the active receptors around them. To improve the function of synthetic receptors, I want to engineer the selective degradation of the inactive receptors by engineering the synthetic receptors to include a degradation tag within them.
What made you interested in pursuing this line of research?
I have always been very interested in learning about the human body and diseases. For a long time, I thought that I wanted to become a doctor, but the more I looked into it, the more I realized that I was more interested in working behind the scenes. Instead of giving the treatments to the patients, I wanted to create the treatments. That’s how I got into the whole realm of chemical engineering and the biological side of chemical engineering.
Why did you apply for the CLP Summer Scholars Program?
I knew that I wanted to continue my research further beyond one or two hours a week during the school year. The CLP program not only provided funding for me to pursue full-time summer research but also introduced me to a small cohort of students with whom I meet regularly. I know that beyond summer, I can continue to connect with this group for the rest of the time I’m here at Northwestern.
What do you like to do when you’re not studying?
I have been playing the piano and the guitar since I was six. For the longest time, I saw that as a huge responsibility. Then, all of a sudden, I had a shift in perspective. Since then, I have enjoyed practicing, especially when I find myself to be stressed or need to take a break from work or studying.

Mychaela Mathews
Mychaela Mathews
Hometown: Vero Beach, FL
Year: 3rd Year
Major: Neuroscience with a concentration in chemistry
CLP Mentor: Yegenia Kozorovitskiy (Neurobiology)
What was the focus of your summer research project?
My lab studies the process of neuroplasticity. Our brain is made up of many different circuits, which is how information is passed from one place to another. These circuits are made up of neurons. Neurons are able to change their appearance and shape through a process called neuroplasticity. There is a chemical called psilocybin that is hypothesized to enhance the growth of protrusions on neurons—called dendritic spines—and therefore encourage neuroplasticity to occur. We expose different intermediates of psilocybin to neurons and analyze how they promote neuroplasticity. This is important clinically because any aberration neuroplasticity can manifest as psychiatric diseases, such as depression and PTSD.
What made you interested in pursuing this line of research?I have always been fascinated by the brain, and particularly the work in Professor Kozorovitskiy’s lab. The idea that we can analyze changes in neurons over time to understand the mechanisms of disease and explore potential treatments is very interesting to me.
Why did you apply for the program?
Eventually, I hope to become a physician. I’m not sure what kind, but I want to be able to elevate the standard of care for my patients by understanding the basic science behind the treatments that are prescribed through research. Although I’ve already had outstanding mentorship in the Kozorovitskiy lab, what has been different about CLP is that I’ve gotten to network with people who aren’t in my lab and have different experiences navigating both medical school and graduate school, which has been extremely helpful as I consider a future career as a physician-scientist.
What do you like to do when you’re not studying?
Time with my friends and family is what I cherish the most. In terms of special interests, I took Swahili for two years at Northwestern, which has been very helpful in enhancing my communication skills, both in Swahili and English. I am also a support group facilitator for a nonprofit called ‘Rainbows For All Children’ based in Evanston which provides mental health resources for children who have experienced loss. I got involved in this organization about a year ago to support others who have also experienced loss. I think it’s important to connect with the community where you live and reflect on how you can use your experiences to positively impact those around you. I also am an avid reader in my spare time.

Adam Suh
Adam Suh
Hometown: Natick, MA/Seoul, South Korea
Year: 4th Year
Major: Chemical Engineering
CLP Mentor: Daniel Arango (Pharmacology)
What was the focus of your summer research project?
I focus on epi-transcriptomic modifications, which are modifications that occur on the RNA, and how those modifications affect translation in cancer cells. Mutant or ‘incorrect’ protein production is one of many characteristics of cancer. We want to answer the question- How does the cell produce these ‘incorrect proteins’ but more specifically, what role do RNA modifications play in mutant protein production? There are many mechanisms known to cause this, but RNA is heavily involved in protein synthesis, so I want to explore its potential. More specifically, my research focuses on a specific modification tied to RNA oxidation, which occurs during a common physiological phenomenon called oxidative stress.
What made you interested in pursuing this line of research?
I have always found molecular biology fascinating. With COVID and the development of mRNA vaccines, there’s been a huge boom in RNA research. RNA is a very dynamic molecule with a lot of roles yet to be discovered. However, because this field is still so new, there are a lot of difficulties with experimental design. I learn something new every single day in the lab and I love being part of something so challenging.
Why did you apply for the CLP Summer Scholars Program?
The CLP is a prestigious grant and my PI suggested that I would be a good candidate for it. I looked more into CLP and talked to two award recipients from last year. I heard great things about CLP faculty and Penelope and the institutional support that goes above and beyond the lab work.
What do you like to do when you’re not studying?
I run a medical student start-up called MedKit. We offer over-the-counter products to undergraduate students on North campus. We are hoping to expand this year, increasing OTC accessibility to other students on campus!
I like a lot of physical activities like running and lifting and playing sports. I ran the Chicagoland marathon about four months ago.
by Lisa La Vallee

Academic, Industry Leaders Gather for the 2023 International Top-Down Proteomics Symposium
One hundred and sixty world leaders in proteomics, proteoform biology, cell biology and genomics gathered October 3-5, 2023, at Northwestern’s Prentice Women’s Hospital to present their latest research findings and discuss next-generation proteomics at the second International Top-Down Proteomics Symposium. The Symposium was hosted by the Consortium for Top-Down Proteomics (CTDP) together with Northwestern University’s Chemistry of Life Processes Institute (CLP).
“Community is a key aspect of science. A strong, cohesive group can catalyze transformative leaps in science and technology – precisely what this community is targeting,” says Northwestern’s Neil L. Kelleher, PhD, Walter and Mary E. Glass Professor of Molecular Biosciences; Professor of Chemistry,; and Professor of Medicine.
Proteins play a central role in maintaining and regulating human health. When proteins confront a challenging environment, disease and illness can result. Scientists are increasingly looking to proteoforms, the exact molecular forms of proteins, as the strongest link between our genes and disease.
“The field of top-down proteomics has advanced tremendously in the last four years. We see new technologies arising and a great future for mass spectrometry to target and tackle proteoforms,” says Julia Chamot-Rooke (Institut Pasteur). “We need to find proteoforms that are associated with disease, and then we’ll be able to develop new clinical assays and understand both the biology of some system organisms, but also how disease progresses and how we can stop it.”
Determination of the precise composition and function of each proteoform within the human body, an approach known as top-down proteomics, enables scientists to identify the true culprits behind disease and develop better targets to eliminate them.
Thirty-six scientific leaders presented and participated in Symposium round tables that covered a diverse range of topics from top-down proteomics approaches and single-molecule sequencing to the clinical value of proteoforms and the advancement of Human Proteoform Project, an ambitious effort to define all of the different forms of proteins in the human body. The Symposium also featured a series of poster and oral presentations covering single-cell methods, proteoform function and complexes, spatial mapping, and clinical applications as well as newer emerging approaches of proteoform analysis such as nanopore protein sequencing, microfluidics, and single molecule protein sequencing.
“You really need to have a few good examples of top-down proteomics that address biological questions to solve clinical problems. That’s what taxpayers want to see. They don’t want to see the numbers or fancy published papers. They want to know—the patient wants to know, ‘Can you treat my disease?’,” says Ying Ge (University of Wisconsin-Madison).
Presenters included Kelleher, the Director of the Chemistry of Life Processes Institute and chair of the TDP2023 Symposium, who shared new approaches to digitizing proteoform biology with single-molecule mass spectrometry. Ben Cravatt (Scripps Research Institute) presented chemical proteomics strategies to discover proteoform-specific small-molecule probes. David Walt (Harvard University) presented hypothesis-driven ultrasensitive methods for the discovery and validation of proteoforms. Amy Herr (University of California-Berkeley and the Chan Zuckerberg Biohub) presented single-cell analysis tools capable of resolving proteoforms and integrating this information into single-cell-omic analysis.

Neil Kelleher welcomes attendees of the 2nd International Top-Down Proteomics Symposium.
The Symposium was generously supported by sponsorships from Bruker, Quantum-Si, SCIEX, Thermo Fisher Scientific, Nautilus Biotechnology, Glyphic Biotechnologies, PacBio, Seer, Agilent, ZefSci, Covaris, CMP Scientific, and Affinisep.
“We are excited to see this level of enthusiasm for advancing the study and impact of proteoforms,” said CTDP CEO Paul Danis. “The momentum from this meeting will propel us forward towards even more wonderful developments and discoveries.”
by Lisa La Vallee

Richard Silverman Receives the 2024 Abeles and Jencks Award for the Chemistry of Biological Processes
Professor Richard B. Silverman of Northwestern University is the recipient of the 2024 Abeles and Jencks Award for the Chemistry of Biological Processes. His first publication with Dr. Abeles (“Inactivation of Pyridoxal Phosphate-Dependent Enzymes by Mono- and Polyhaloalanines”) set the stage for a truly distinguished career and a body of work that showed how mechanistic enzymology could be used to develop a drug by unravelling the fine details of the inactivation process and using these findings to make efficient and specific inactivators. PLP-dependent enzymes were long thought to be “undruggable” because so many enzymes use PLP that one could never achieve specificity, resulting in a host of adverse effects. Rick’s work is a model for the bench to bedside development of mechanism-based inhibitors of PLP-dependent enzymes. Among his many accomplishments, he invented Lyrica, the blockbuster drug marketed by Pfizer for epilepsy, fibromyalgia, and neuropathic pain. More are in the pipeline or in clinic trials.
Rick has been at Northwestern University since 1976. The field of enzymology has been bestowed with outstanding scientific talent. Those investigators who have not only elucidated the chemical mechanism of an enzyme but have rationally-designed molecules that cleverly exploit an enzyme’s mechanism to activate a pseudo-substrate that goes on to covalently inactivate the enzyme, known as mechanism-based inactivation, stand at the top of this field of study. Rick literally wrote the book on mechanism-based inactivators. Fittingly, Rick began studies on mechanism-based enzyme inactivation in the laboratory of Dr. Abeles. Professor Silverman is one of the stars from this legendary laboratory.
Story first published on October 9, 2023, by The ACS Division of Biological Chemistry.

CLP Receives $2M to Prepare the Next Generation of Science Innovators
The Chemistry of Life Processes Institute (CLP) recently received a $2 million five-year National Institutes of Health (NIH) T32 training grant award (1 T32 GM 149439-01) to prepare a diverse cohort of students to lead the next wave of innovation and discovery at the interface of chemistry and biology.
The award follows ten years of NIH/NIGMS T32 funding in support of CLP’s highly regarded Predoctoral Training Program. In addition to providing evidence-based training and mentoring approaches that incorporate coursework, research experience, and career development, the new award requires funded institutions to ensure diversity at all levels—from the science topics covered to the recruited students.
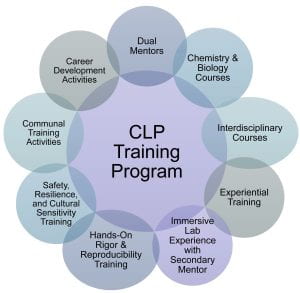
Features of the CLP Predoctoral Training Program
Over the past decade, 26 graduate students have completed the CLP Predoctoral Training Program and have gone on to pursue successful careers in academia, industry and government. The Training Program provides students with the opportunity to integrate graduate studies in chemistry and the life sciences and become immersed in the highly collaborative research programs of 52 mentors from 3 schools (Weinberg, Feinberg and McCormick) across 10 departments.
The depth and breadth of the CLP Program coursework requirements, novel immersive cross-disciplinary lab experience, and unique mentoring structure, coupled with a strong shared training environment, sets it apart from other biomedical training programs,” says Sheila Judge, CLP’s Senior Director for Research, Education and Administration.
The Training Program also supports trainee career development, communication skills training, resume development, and leadership skills. Activities such as improvisational communications workshops led by an instructor with Chicago’s The Second City, as well as workshops with industry leaders and invited talks by investigators from other academic institutions expose trainees to alternative career pathways and facilitate networking with potential mentors.
The new award funds eight trainees per year. Trainees are appointed to the grant starting in the fall of their second year and are supported throughout their second and third years of graduate education. Two additional students are supported annually by one-year university fellowships, enabling the participation of well-qualified and diverse international students.
The CLP Training Program deploys a team-based approach to program leadership that leverages the unique expertise of its members and the administrative capabilities of the CLP to enhance program development, assessment and evaluation, and training. The team is led by PI/Program Director Neil Kelleher, who is assisted by three Associate Directors, Regan Thomson (Chemistry), Amy Rosenzweig (Molecular Biosciences), and Sheila Judge (CLP), and program administrator Penelope Johnson.
“The new evidence-based training methods and approaches developed by the CLP program will be shared with graduate programs at Northwestern and across the country,” says Judge. “Our goal is to maximize the impact of program innovations and contribute to the development of 21st-century graduate STEM education.”

New CLP summer research program trains and mentors NEIU undergraduates
This summer, Northwestern’s Chemistry of Life Processes Institute (CLP) welcomed the first participants of its Interdisciplinary Summer Undergraduate Research Experience (I-SURE), a new partnership with Northeastern Illinois University (NEIU). The eight-week summer program provides promising undergraduates from the federally designated Hispanic-Serving Institution with interdisciplinary research training and mentorship in CLP faculty laboratories.
“In keeping with Northwestern’s commitment to diversity and inclusion, I-SURE enables students from underrepresented communities in STEM to gain research skills and confidence to pursue careers in biology, chemistry, engineering, technology and medicine,” says Sheila Judge, CLP’s Senior Director for Research, Education and Administration. “The program also provides a bridge to Northwestern graduate programs, as well as the CLP Predoctoral Training Program. We are delighted to support two outstanding students, Gabriel Urbina and Abigail (Abi) Lopez Alonso, in the inaugural cohort for this program.”

Gabriel Urbina, NEIU biology major
One semester short of completing his bachelor’s degree in math at NEIU, Gabriel Urbina put his education on hold in 2015 to rethink his academic and career goals. In addition to math, he was equally passionate about art and science. At one point, he seriously considered moving to New York to become a full-time painter but his interest in science prevailed.
“During the pandemic, I used to read a lot of scientific articles and listen to podcasts related to science, particularly microbiology and how fast microorganisms evolve and how they’re able to adapt to so many environments,” says Urbina. “It sounds like some kind of dystopian science fiction, but it’s becoming a reality—hospitals are having problems with antibiotic-resistant and antiseptic-resistant bacteria. It’s a double-edged sword because if you’re in a hospital for a long time with cancer, for instance, you have a higher chance of catching these mutated bacteria and infections and that is a big killer for these people.”
Last year, Urbina completed his math degree and immediately began a second bachelor’s degree in biology. He applied for and was accepted into CLP’s I-SURE program and is working in the lab of CLP member Erica Hartmann (biomedical engineering). His research project focuses on hospital bacteria that are resistant to cleaning chemicals and how they evolve and pass on their genes.
Urbina hopes to attend graduate school within the next few years and pursue a career in academia.

Abigail Lopez Alonso, NEIU chemistry major
Before applying to the I-SURE program, Abi Lopez Alonso worked as a medical assistant in various clinics and hospitals. In her last position, she was in charge of dropping off blood samples at the lab, a job that intrigued her.
“I asked, what do you do to get here? A lot of them told me that they had a degree in chemistry,” she says.
Lopez Alonso went back to school intending to become a lab technician, but her professors encouraged her to look into a PhD, an idea she initially believed was “too advanced” for her. After receiving third place for her summer research project at Northeastern last year, she began to reconsider.
“I really wanted to get more lab experience outside of school and see what a PhD program is like from the student’s perspective,” says Lopez Alonso. “Right now, I am paired up with a PhD student [in the lab of Danielle Tullman-Ercek (Chemical and Biological Engineering)], so I get to ask a lot of one-on-one questions and see the day-to-day to make sure it’s something I actually want to do before I jump in.”
The I-SURE trainee says that she is looking forward to gaining hands-on lab experience doing research with promising pharmaceutical applications.
“Going to school is one thing in that you learn all the theory, but this is really exciting because it is all cutting-edge stuff that’s not in textbooks and hardly in the news,” says Alonso. “Learning the real lab techniques of how to do things is the most exciting part for me.”
I-SURE builds on over a decade of CLP experience managing summer research programs for Northwestern undergraduates. The program provides a stipend, lab expenses, and an opportunity for students to present at the annual Chicago Area Undergraduate Research Symposium. The 2023 program was made possible through the generosity of several members of the Institute’s Executive Advisory Board.
To identify potential candidates. CLP partners with the NEIU Student Center for Science Engagement.
“I am incredibly excited about this new connection between Northwestern and NEIU,” says Jorge Cantu, assistant professor of biology, and director of NEIU’s Student Center for STEM Engagement. “The faculty at NEIU and the Student Center for Science Engagement has nurtured a robust undergraduate research community that seeks opportunities like I-SURE to showcase their incredible talent and potential as scientists.”
Preparing a talented pool of underrepresented researchers, Cantu points out, is still only half of the equation.
“Regrettably, summer research programs are often underprepared to accommodate our best candidates,” Cantu says. “This is not the case with the I-SURE program.
“The CLP program was intentional in seeking diverse voices in their mentoring networks and tailoring support systems for the unique needs of our students, who are non-traditional, first-generation, and trained at a primarily undergraduate institution. I cannot wait to open this opportunity to more of our NEIU students in the coming years.”
by Lisa La Vallee
Pioneering New Methods to Understand Protein Folding
Northwestern Medicine scientists have developed a new technique for measuring protein folding stability on an unprecedented scale, findings detailed in a new study published in Nature.

Gabriel Rocklin, PhD, assistant professor of Pharmacology, was senior author of the study published in Nature.
While advances in technology have helped scientists discover new protein sequences and folded structures, the overall stability of these folded structures are still largely a mystery and new methods are needed to reveal these folding behaviors.
Better understanding of protein folding may provide insight into disease development and protein evolution, as well as guide future approaches to protein engineering and drug development, said Gabriel Rocklin, PhD, assistant professor of Pharmacology and senior author of the study.
“What’s significant about our paper is that we can use this approach to measure folding stability for practically a million different sequences in each experiment,” Rocklin said. “Measuring stability at that scale has always been impossible and a big bottleneck for research. Until recently, the main way scientists measure protein stability is purifying one protein at a time and doing an experiment on that one protein. With individual measurements like this, it’s hard to make predictions about stability for new sequences. By measuring stability on this much larger scale, our data can be used to develop machine learning tools to predict stability and design higher stability proteins.”
In the study, scientists combined the strengths of cell-free molecular biology and next-generation sequencing to develop a new method—dubbed cDNA display proteolysis—to produce a dataset of 776,298 protein folding stability measurements.
Compared to other high-throughput stability assays such as mass spectrometry, the cDNA proteolysis approach produces 100 times more data on protein folding, making it a useful dataset for training machine learning algorithms for experiments involving protein design in the future, according to the authors.
The dataset produced is also unique in that it covers all single protein mutants for 479 different protein domains, as well as more than 200,000 double mutations that provide additional insights into folding. This scale and systematic design make it one of the most comprehensive sources of data of its kind currently available to the scientific community.
“Folding stability is this really important property because it influences function and aggregation and whether a designed protein is going to be a useful therapeutic or not,” Rocklin said. “And if we want to be able to predict that quantity, we need data on this scale. This method that we introduce is the first approach that is going to get us the stability data on the scale that we need for machine learning.”
Moving forward, Rocklin and his collaborators will use the dataset to train machine learning models and use those models for protein engineering, he said.
“What we are working on now is taking the data that we already have and creating a machine learning model with it,” Rocklin said. “We’d also like to take the experimental approach that we have and expand it to all types of protein domains, especially even larger proteins. We’ve already had a lot of interest from other scientists in using our dataset, so we’re excited to empower computational researchers and biophysicists and accelerate machine learning in protein science.”
Kotaro Tsuboyama, a former postdoctoral fellow in the Rocklin lab and currently a lecturer at the University of Tokyo, was lead author of the study.
Rocklin is a member of Northwestern’s Center for Synthetic Biology, Chemistry of Life Processes Institute, and Robert H. Lurie Comprehensive Cancer Center of Northwestern University. The cell-free approaches employed in the study are one of the research specialties of the Center for Synthetic Biology.
The study was supported by Northwestern University Startup Funding, Japan Society for the Promotion of Science grant 19J30003, the Human Frontier Science Program Long-Term Fellowship, and Japan Science and Technology Agency grant JPMJPR21E9.
The original story by Olivia Dimmer was published on July 19, 2023, in Feinberg News.
